Sim2Virus User's Manual
Overview:
Sim2Virus is a Java applet that models the competitive spread of two types
of virus on a hexagonal grid of cells. Infected cells are colored red or green
according to the type of virus, with uninfected cells remaining white. In the
case that simultaneous infection of a cell by both types of virus is allowed
(superinfection), an intermediate color is used, with the median value being
displayed as yellow. When a cell dies it retains its final color.
Initial infection:
The intial infection occurs in areas called the foci, one for each type of virus.
The default is that both foci are in the center of the screen and are the same
shape, so they overlap exactly, and that both infections start simultaneouly.
If a nonzero value is entered for the DeltaX parameter the green focus
is moved to the left by that number of cells and the red focus is moved
to the right by the same distance. If a nonzero value is entered for the
DeltaT parameter the red infection starts later by that number of time steps.
The user can also specify the radius of each focus and select from one of four
possible shapes:
- Hexagon - a regular hexagon
- Hex2 - a hexagon with one vertex moved to the center
- Hex3 - Hex2 with the opposite vertex moved to the center as well.
This gives two triangles connected at a vertex.
- Disc - a circular disc.
The number of viruses of each type in the initial infection is controlled
by the MOI parameters. The MOI specifies how many of each type of virus
is placed on the focus. The number of cells in the focus is multiplied
by the MOI, and this quantity of virus as spread uniformly at random
across the focus. Any cell that gets a receptor in the first time step
will become infected with the viruses on that cell. Receptors are
assigned at random at a rate given by the "% infectible" field.
The user can also turn on a thin black line delineating the border of the
focal area. This is purely a visual aid - it has no effect on the simulation.
Cell life cycle:
The following diagram illustrates the life-cycle of a cell:
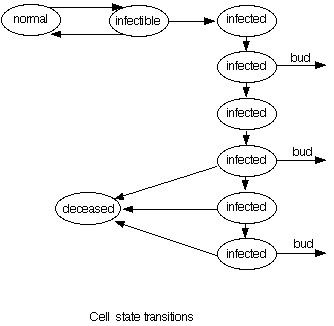
The six states labelled "infected" represent the fact that a cell lives for
at most six time steps before it dies. This is controlled by a timer
that is set to a random value of 4, 5, or 6 when the cell first becomes
infected. Viruses bud out of infected cells only from the second and
fourth (and possibly sixth) of these states.
State transitions:
- normal to infectible
- This transition models a cell acquiring a receptor. At each step (including
the initialization step), a fraction of cells given by the "% infectible" parameter
acquires a receptor. For each cell that acquires a receptor, a timer is set
to a uniform random value ranging from "Min RRT" through "Max RRT"
(Receptor Retention Time).
- infectible to normal
- If the above timer expires before the cell becomes infected it returns to normal
- infectible to infected
- This transition models a cell becoming infected when virus is present at the
same time the cell has a receptor. This can happen in the same time
step as the cell acquires the receptor (normal to infectible transition).
At this point, the cell's timer is set to a uniform random value between
4 and 6 inclusive. See "Virus lifetime" (below) for an explanation of
how viruses enter a cell.
- infected to infected
- The "infected" state is made up of six substates. The number of
states traversed is determined by the setting of the cell's timer when
it enters the first "infected" state. Viruses bud out from the cell
to the six adjacent cells at the second, fourth, and (if the timer was
set to six) sixth states. When budding occurs, an average of about
100 viruses are distributed randomly to the six adjacent cells. If the
adjacent cell is already infected and superinfection is allowed the
viruses enter the cell directly, otherwise they wait on the surface
of the cell. If the budding cell was infected by both types of virus
they are distributed in proportion to the ratio in that cell.
- infected to deceased
- A cell remains infected for between 4 and 6 time steps, after which it dies.
Virus lifetime:
The length of time a virus can live outside a cell is controlled by the
"Max age" parameter. Allowable values are from one to four time steps.
When a cell becomes infectible, the number of live viruses on the surface
of the cell is calculated. If superinfection is not allowed, the cell becomes
infected by the virus that is in the majority. If superinfection is allowed,
viruses enter the cell in proportion to the ratio on the surface, however
there is also a probability that the cell becomes infected with one type
of virus. This probability increases linearly from 0 (if the two types of
virus occur in equal numbers) to 1 (if only one type of virus is present).
Applet controls:
The layour of the applet looks like this:

- MOI
- The "MOI" fields allow the user to specify (on average) how many viruses of each type are placed on cells in the focus initially
- Max age (choice menus)
- The number of time steps a virus can live outside a cell
- Focus shape (choice menu)
- The shape of the area where the initial viruses are deposited (see "Initial infection, above)
- Focus radius
- The value in this field specifies the size of each focal area
- Superinfection (radio buttons)
- If superinfection is allowed a cell can become infected by both types of virus simultaneously
- Green only
- Show only the green viruses. This affects only the display - the red viruses are still present in the simulation
- Red + Green
- Show both types of virus
- Red only
- Show only the red viruses. This affects only the display - the green viruses are still present in the simulation
- Border on/off
- Displays or hides a thin black outline around the focus
- Restart
- Restart the simulation with a new initial infection
- Run
- Continue simulating the current infection
- DeltaX
- The center of the green focus will be moved left by this number of cells,
while the center of the red focus will be moved right by the same number.
- DeltaT
- The time step when the initial infection for the red virus should start.
The initial infection for the green virus always starts at time 0.
- % infectible
- The proportion of cells becoming infectible (acquiring a receptor) at each time step
- Min RRT
- Minimum Receptor Retention Time, the minimum number of time steps
that a cell remains infectible
- Max RRT
- Maximum Receptor Retention Time, the maximum number of time steps
that a cell remains infectible
- Iterations
- The number in this field specifies how many time steps are simulated with each click of the "Run" button
Statistics window:
This window displays statistics of the numbers of
cells with different ratios of red and green viruses.
The leftmost column is the time and the next column is the number of
cells that are all green. The rightmost column is the number of cells
that are all red. The remaining columns are ten percentile counts
for cells that have a mix of red and green.
A view of the statistics window after 30 time steps have been simulated:




