Sample publications:

|
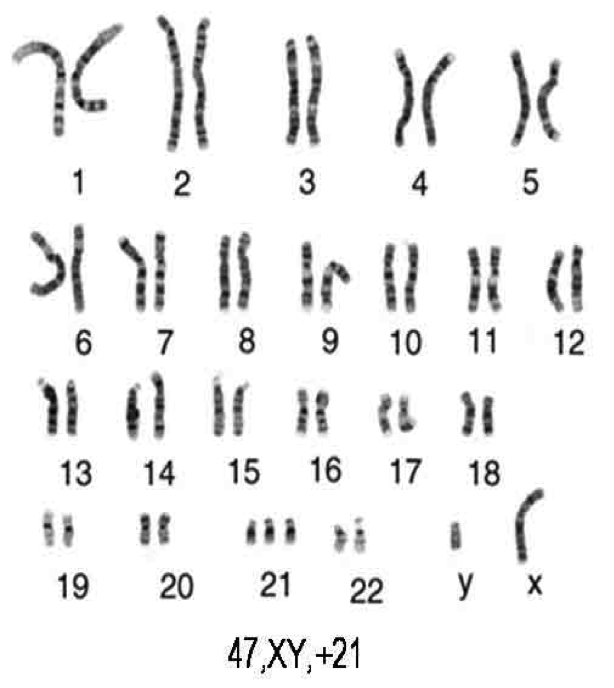
We are interested in what can be learned about fetal and neonatal health
from transcriptional profiles, and we have performed a number of studies profiling expression
in maternal or neonatal peripheral blood, amniotic fluid, and cord blood, both under
normal conditions and in the presence of developmental anomalies.
Our early work in this area
demonstrated that the limited gene functional annotation in this domain made it
challenging to interpret such studies. Thus, we have also worked to improve the
annotation infrastructure relating to human development, contributing annotations to the Gene
Ontology and to DFLAT, a collection of expanded gene sets within the GO framework,
tailored to support gene set analyses of developmental transcriptomes. We are
currently applying molecular methods to predict complications in NICU patients
and to understand the neurodevelopmental and metabolic impacts of maternal opioid use.
Sample publications:
|
| In the era of precision medicine, we need techniques for
identifying significant and meaningful characteristics of individual
patients given high dimensional genomic data such as
gene expression or sequence variation. We have pioneered new
computational approaches to anomaly detection (the analogous machine
learning challenge), and applied these methods to find functional
pathways whose anomalous regulatory expression patterns characterize
individual anomalous samples. Such methods are particularly useful
when individual anomalous samples are known to be rare unique
anomalies, or when multiple samples are typically classed together by symptoms
but thought to be caused by heterogeneous molecular processes.
Sample publications:
|
|
|
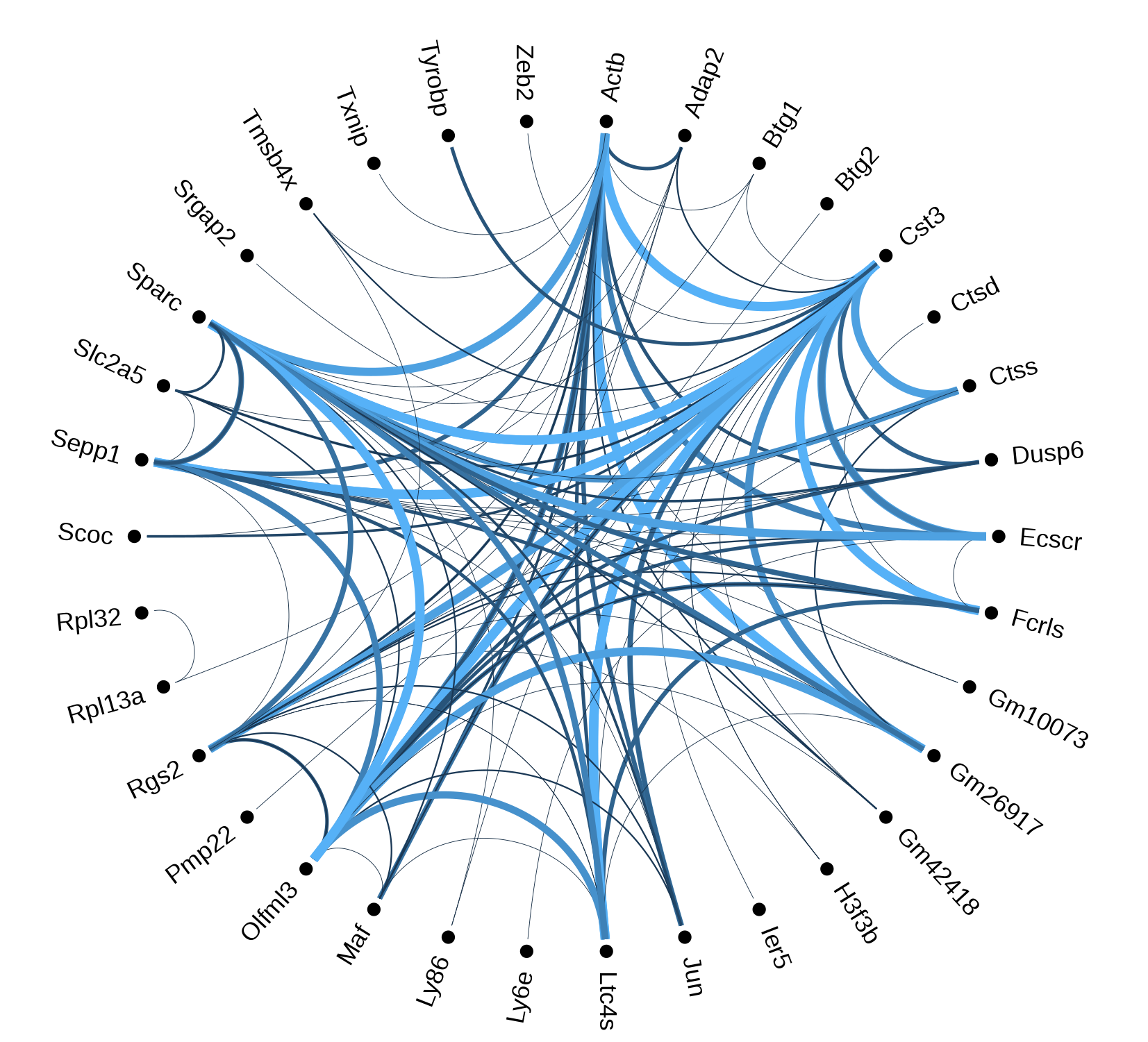
One of our key interests is understanding more about how genes
and proteins interact to carry out the molecular functions needed
by living organisms. We are interested in understanding network
properties, using these properties to infer functional roles of
genes and their relationships to disease, and
adding biological context to reported interactions.
Studying the connectivity and temporal properties of biological
networks may help us design better diagnostics and therapeutics,
identify novel drug targets, and understand broad properties of
cellular behavior and evolution. We are also interested in using
computational approaches to discover new disease genes and the roles
that functional processes or pathways play in disease processes.
Sample publications:
|
|
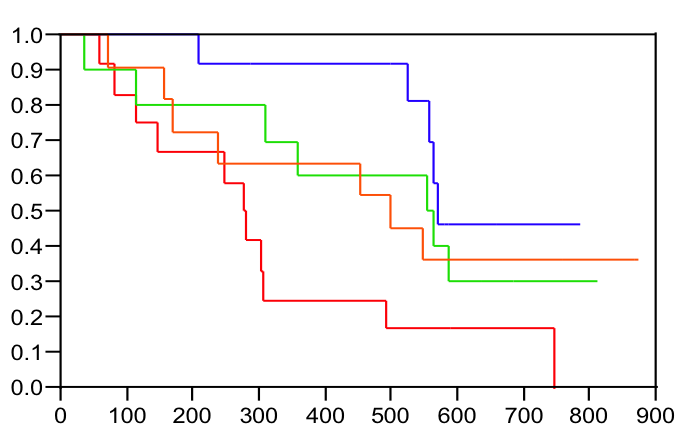
Pharmacogenomics - the use of genomic techniques to develop better
drugs to treat disease - remains an exciting research area for
bioinformatics scientists. I've long been interested in developing predictive
models of drug response to help tailor medications to those
individuals most likely to benefit or to identify individuals at
highest risk of adverse side effects. I'm also interested in using
bioinformatics to help identify novel indications for existing
compounds.
In the future, developing new methods to identify better
drug targets has the potential to revolutionize the pharmaceutical
industry, leading to better understanding of mechanisms of action,
more personalized treatments, and lower costs for both producers and
consumers.
Sample publications:
|
|