|
The mission of my research lab is to develop
ANALYSIS + DESIGN
tools to advance (RE-)DESIGNING BIOLOGY. Our tools provide insight into complex biological systems. They also enable building novel biological components to produce useful chemicals and therapeutics. My lab now focuses on developing
MACHINE LEARNING
models that are custom-tailored for biological data to build such tools. My industrial and academic experiences in design automation for electronic systems informs how I approach re-designing biology.
|
|
|
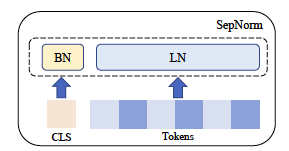
Separate normalization of normal and [CLS] tokens in self-supervised transformers.
Transformer models typically utilize a single normalization layer for both the class token [CLS] and normal tokens. We show a 2.7% performance improvement in image, natural, and graph tasks when utilizing separate normalization layers.
|
|
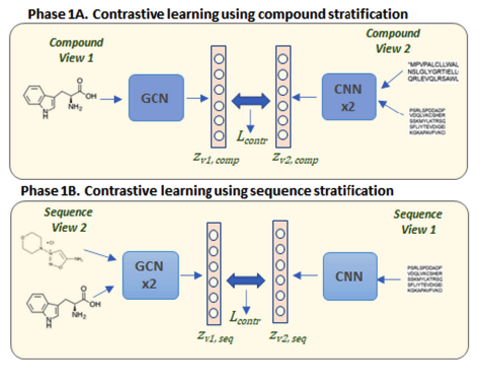
Enzyme-substrate interaction prediction using contrastive multiview coding (CMC).
Two enzymes and a molecule; two molecules and an enzyme.
These are two different views of an interaction. We stratify data and use CMC to outperform all known interaction prediction methods.
|

|
What's new in computational methods for Metabolomics?
Review article on novel methods for mass spec analysis with colleauges from the 2022 Dagstuhl seminar, “Computational Metabolomics: From Spectra to
Knowledge”.
|
|
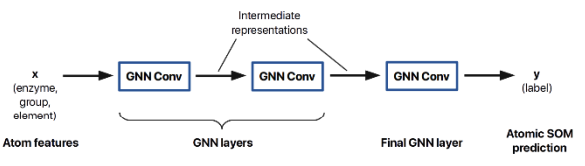
Predicting site-of-metabolism for enzymatic reactions.
Molecules are graphs. We can use GNNs to build SOM predictors for all classes of enzymes! Results demonstrate benefits to enhancing rule-based biotransformation prediction methods.
|
|
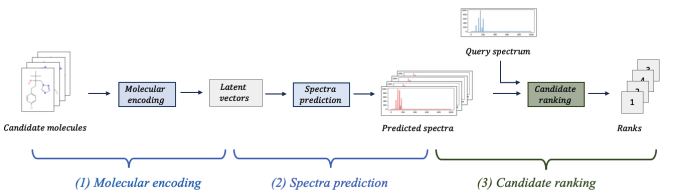
Learning to rank for metabolite annotation for metabolomics.
Ensemble Spectral Prediction (ESP) impelments a novel pipleine that includes: molecular representation learning, spectral prediction with peak co-dependency analysis, and rank-based learning.
Performance is improved up to 41% over MLPs.
|
|
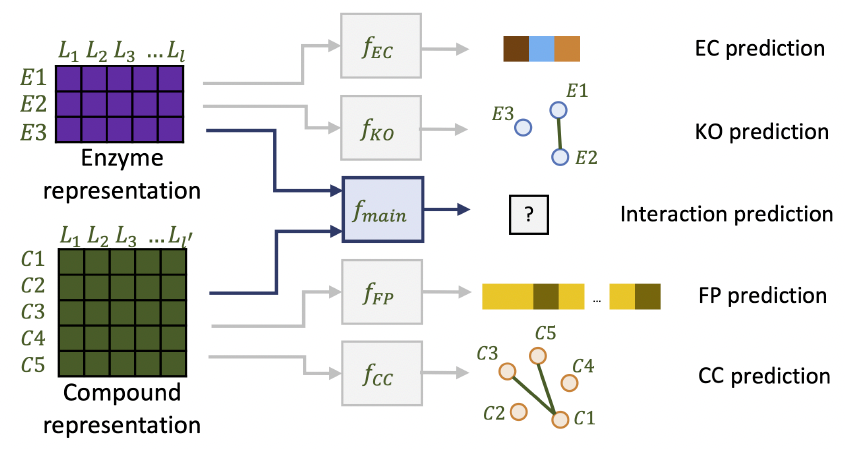
Recommender Systems to recommend enzymes to molecules and vice versa.
Boost-RS is a general recommender system that utilizes relational and group auxiliary data to boost learned
representations. Boost-RS's versatility is demonstrated for matching molecules and enzymes.
|
|
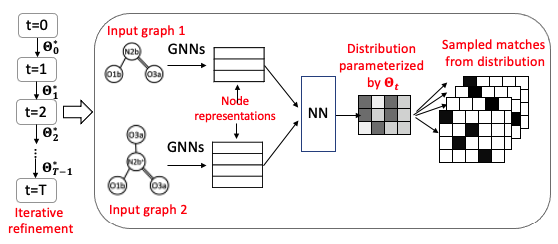
Graph Matching and molecular alignment. A novel deep learning method for aligning two graphs,
with application to molecular matching. Useful for knowing where two molecules match and differ.
|
|
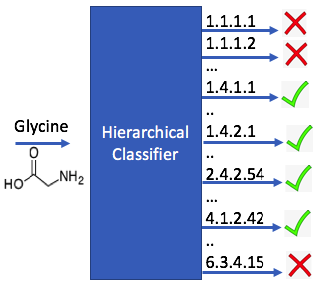
Enzyme Classification on Molecules. A new method for training enzyme-specific predictors that take as input a given query substrate molecule and return whether the enzyme would act on that substrate or not.
|
|
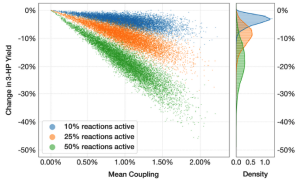
Metabolic Disruption analysis in engineered hosts. Engineering cellular hosts may result in unexpected and undesirable byproducts due to promiscuous interactions of native/heterelogous enzymes and molecules. These effects are not only disruptive to the host metabolism but also to the intended end-objective of high yield. How do you analyze such disruptions? MDFlow is the answer!
|
|
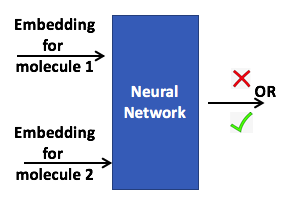
Enzymatic Link Prediction. Learning graph representations of biochemical
networks and its application to predicting enzymatic links between two molecules.
|
|
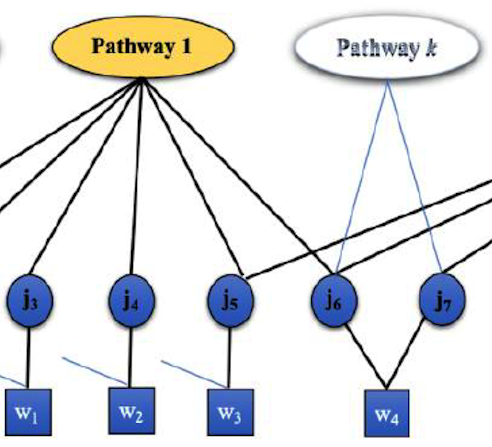
Probabilistic Analysis of Pathway Activities and Metabolite Annotations. Using inference, we learn the likelihood of metabolic pathways being responsible for presence of metabolomics measurements, and the likelihood of annotations.
|
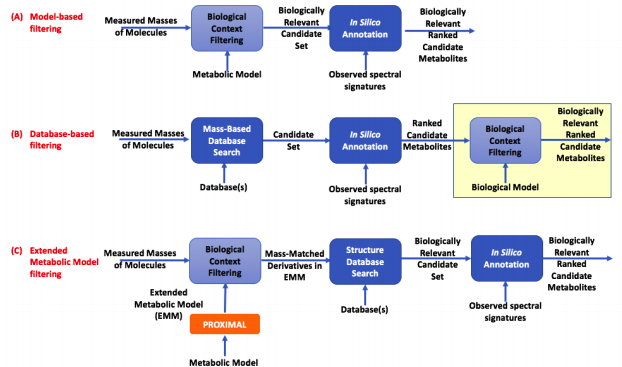 |
Extended Metabolic Models (EMMs). This workflow utilizes our tool PROXIMAL to create Extended Metabolic Models (EMMs) that contain not only canonical substrates and products of enzymes already cataloged for an organism, but also metabolites that can form due to substrate promiscuity. We created an EMM for E. coli and to analyze metabolomics data for CHO and murine cecal microbiota.
|
|
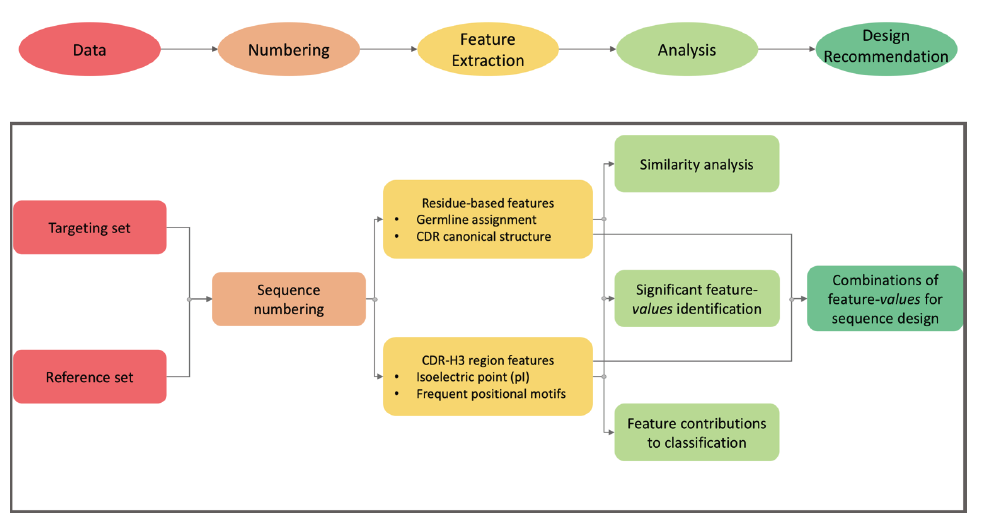
Machine learning analysis pipeline for antibody sequences. Important features in antibody sequences are identified relative to a reference set using machine learning and statistical analysis.
|
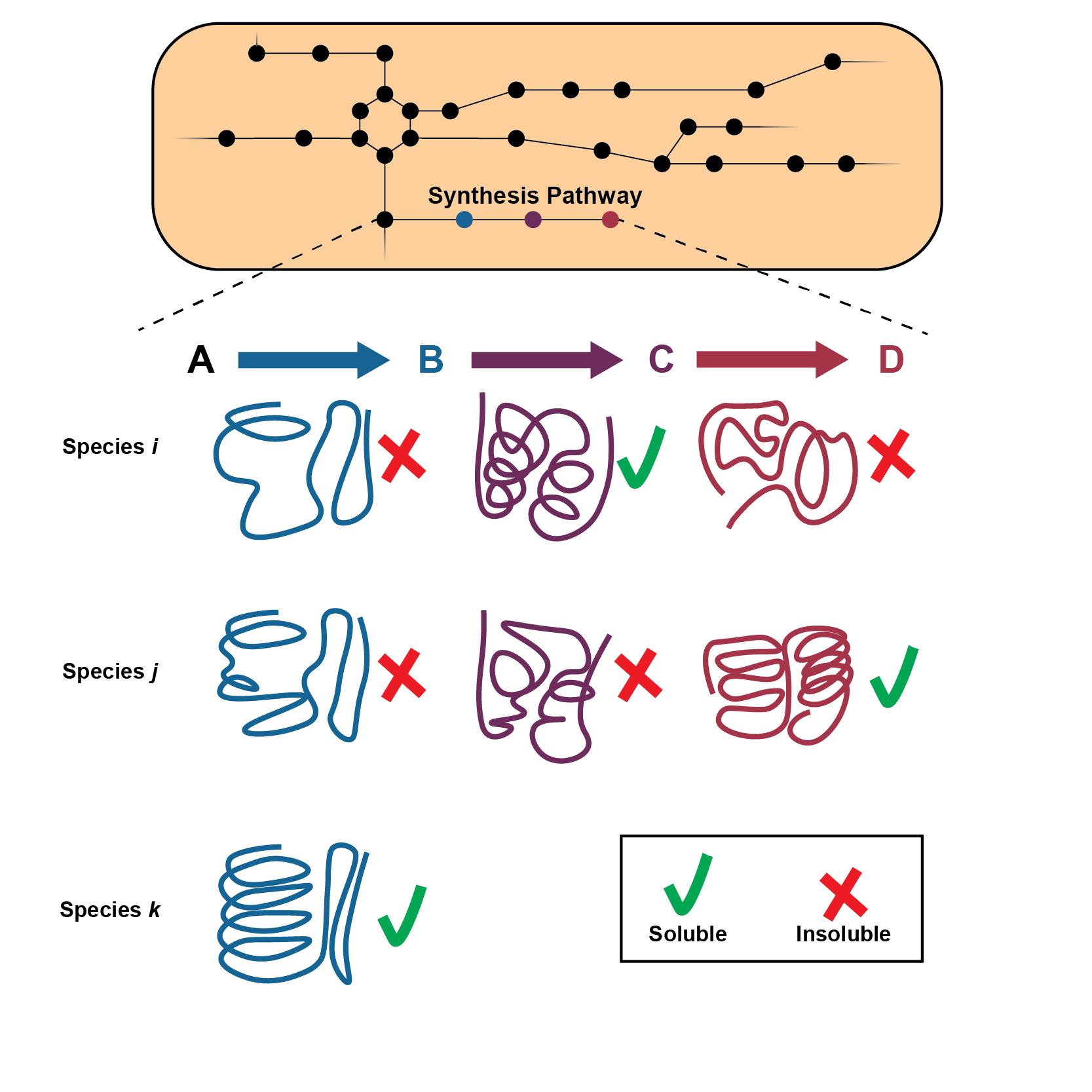
|
ProPASS. A workflow that links synthesis pathway construction with the exploration of available enzyme sequences that are predicted soluble in the host. ProSol DB is a database cataloging the predicted solubility of over 250,000 reviewed enzyme sequences from UNIPROT.
|
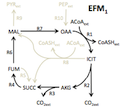 |
gEFM. A method for computing elementary flux modes.
|

|
PROXIMAL. A method to derive biotransformation operators from a reaction and to apply them to a target molecule. Operators can be applied to predict xenobiotic products (as suggested in the paper), or applied more generically to identify derivative metabolites for a specific query molecule and a specified enzyme.
|
















